|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2013 | Volume
: 19
| Issue : 4 | Page : 392-396 |
| |
Genetic variants in the cytochrome P450 2D6 gene in the Sri Lankan population
T. D. Praveen Tharanga1, C. M. V. Jinadasa2, MF Risama2, Priyadarshani Galappatthy1, RL Jayakody1, Vajira H. W. Dissanayake2
1 Department of Pharmacology, Faculty of Medicine, University of Colombo, Colombo 00800, Sri Lanka
2 Department of Human Genetics Unit, Faculty of Medicine, University of Colombo, Colombo 00800, Sri Lanka
| Date of Web Publication | 4-Jan-2014 |
Correspondence Address:
Vajira H. W. Dissanayake
Human Genetics Unit, Faculty of Medicine, University of Colombo, Kynsey Road, Colombo 00800
Sri Lanka
 Source of Support: By a Grant from them Improving Relevance and
Quality of Undergraduate Education Project, Faculty of Medicine, University
of Colombo, Sri Lanka, Conflict of Interest: None  | 3 |
DOI: 10.4103/0971-6866.124361

 Abstract Abstract | | |
Introduction: Cytochrome P450 2D6 (CYP2D6) enzymes are involved in the metabolism of a large number of commonly prescribed drugs such as antidepressants and cardiovascular drugs. The CYP2D6 *3, *4 and *14 variants associated with the loss of enzyme function; CYP2D6 *10 and *17 variants with reduced enzyme function; and CYP2D6 *2 variant with no effect on enzyme function. Establishing the frequency of these variant alleles in Sri Lankan population would be useful for optimizing pharmacotherapy with CYP2D6-substrate drugs.
Objective: The objective of this study was to determine the prevalence of CYP2D6 *2, *3, *4, *10, *14 and *17 variants in the main ethnic groups in the Sri Lankan population.
Materials and Methods: A total of 90 deoxyribonucleic acid (DNA) samples (30 each from Sinhalese, Tamils and Moors) were selected from a DNA resource at the Human Genetic Unit, Faculty of Medicine, University of Colombo. This collection had been made for population genetic studies from a random population based volunteers. Genotyping was performed using published polymerase chain reaction/restriction fragment length polymorphism methods.
Results: The prevalence of the CYP2D6 variants in Sinhalese, Sri Lankan Tamils and Moors respectively were CYP2D6 *2: 37%, 41.6% and 37.9%; CYP2D6 *3: 60.3%, 45% and 30%; CYP2D6 *4: 21.6%, 6.6% and 8.3%; CYP2D6 *10: 40%, 35% and 44%. CYP2D6 *14 and *17 variants were not identified.
Conclusion: CYP2D6 *3, *4 and *10 variants, which are associated with reduced or loss of CYP2D6 enzyme function were found in our population in significant frequencies. CYP2D6*4, which is reported to be a Caucasian variant was also found in all three ethnic groups.
Keywords: Allele frequency, cytochrome P450 2D6, Sri Lankan
How to cite this article:
Tharanga TP, Jinadasa C, Risama M F, Galappatthy P, Jayakody R L, Dissanayake VH. Genetic variants in the cytochrome P450 2D6 gene in the Sri Lankan population. Indian J Hum Genet 2013;19:392-6 |
How to cite this URL:
Tharanga TP, Jinadasa C, Risama M F, Galappatthy P, Jayakody R L, Dissanayake VH. Genetic variants in the cytochrome P450 2D6 gene in the Sri Lankan population. Indian J Hum Genet [serial online] 2013 [cited 2016 May 24];19:392-6. Available from: http://www.ijhg.com/text.asp?2013/19/4/392/124361 |
 Introduction Introduction | |  |
The cytochrome P450 family of enzymes plays a vital role in the metabolism of both endogens and exogenous molecules. Inter-individual variability in drug response and toxicity is accounted for by the variability in the activity of these enzymes. [1] Cytochrome P450 2D6 (CYP2D6) is a member of this family of enzymes. Over 50 drugs, including antiarrhythmics, antidepressants, β-adrenoceptor blockers, neuroleptics and opioids have been identified as substrates of CYP2D6. [1] Seventy four allelic variants and sub-variants have been reported in the CYP2D6 gene and the number is growing. [2] The genetic variations of alleles are classified into groups according to their enzyme activity. The enzymatic activity of the common alleles is as follows: Alleles *1 and *2 do not alter the activity of the enzyme. [3] Alleles *10, *17, *36 and *41 result in an enzyme, which has partially lost its enzymatic activity. [2],[3] Alleles 3, *4, *5, *6, *7, *8, *11, *12, *13 and *14, (called null alleles result in an enzyme, which has completely lost its activity. [2],[3] The clinical importance of the alleles is being increasingly recognized as they are known to cause alterations in drug clearance, response to drugs and side-effects.
The activity level of the CYP2D6 enzyme depends upon the combination of CYP2D6 alleles present in an individual. Depending on the activity of the CYP2D6 enzyme individuals can be classified as poor metabolizers (PM), intermediate metabolizers (IM), extensive metabolizers (EM) and ultra-extensive metabolizers (UM). The PM phenotype results when an individual has two null alleles. The IM phenotype results in three ways, i.e. presence of two partial loss alleles, presence of one null allele and one partial loss allele or presence of one null allele and one normal allele. The EM phenotype results when an individual has two normal alleles. The UM phenotype results when there are more than two normal alleles, which can occur due to amplification of the CYP2D6 gene. [3]
It is being increasingly recognized that dosage recommendations for the above mentioned classes of drugs, avoidance of side-effects and therapeutic failure, can be made based on the CYP2D6 genotype. [4],[5] On one extreme, PM have a higher risk of developing concentration-related side-effects and toxicity. On the other extreme, EMs is likely to experience therapeutic failure with drugs such as nortriptyline, tramadol, morphine and metoprolol. [1],[3] The functional impact of CYP2D6 alleles is known to be substrate dependent. [3] For example CYP2D6 *17, is generally considered as an allele, which reduces function, but it displays remarkable variability in its activity toward substrates such as dextromethorphan, risperidone, codeine and haloperidol. [3]
The distribution of these alleles in various world populations is not uniform. Some of these alleles are reported to be population specific: allele *10 is found in Asians; [6] allele *17 is found in Africans; [7] and allele *4 is found in Caucasians. [8] A previous study in a Sinhalese population in Sri Lanka, in the early 1990s, reported on the frequency of a limited number of variations in the CYP2D6 gene. [4] Since then, knowledge in this field, including the nomenclature used to refer to the variations has advanced greatly and a fresh examination of the prevalence of these variations in Sri Lanka would be timely. The objective of this study, therefore, was to determine the prevalence of the CYP2D6 *2, *3, *4, *10, *14 and *17 alleles in Sinhalese, Sri Lankan Tamils and the Moor in the Sri Lankan population.
 Materials and Methods Materials and Methods | |  |
Subjects
An already established population based deoxyribonucleic acid (DNA) resource consisting of 30 each of Sinhalese, Sri Lankan Tamil and Moor volunteers (50% male) was used for this study. This population based DNA resource is maintained at the Human Genetics Unit, Faculty of Medicine, University of Colombo. This collection had been made with the approval of the Ethics Review Committee of the Faculty of Medicine, University of Colombo for population genetic studies of this nature after obtaining written informed consent from the volunteers. Ethics approval was obtained from the same committee to use already stored DNA samples for this study.
Genotyping methods
Genotyping was carried out using allele specific polymerase chain reaction (PCR) or PCR-restriction fragment length polymorphism (PCR-RFLP) methods. PCR mixture (25 μl) was prepared using 30-40 ng of DNA, 2.5 μl of 10* PCR reaction buffer, 1.5 μl of 25 mM MgCl 2 , 2.5 μl of dNTP, each of the respective primers at 20 pmol/μl, 2U of Taq polymerase (Genei). Amplification was checked on 1% agarose gel at 60V for 10 min. In PCR-RFLP protocols, the amplified PCR products were digested using appropriate restriction enzymes and the digested PCR products were separated by electrophoresis using 2% agarose gel at 60V for 40 min. DNA bands were visualized by staining with ethidium bromide. Genotypes were determined the based on the size of the respective DNA fragments.
Statistical analysis
Allele frequencies were calculated and tested for Hardy-Weinberg equilibrium. Chi-square test was used to compare allele frequencies between populations. P < 0.05 was considered to be significant.
 Results Results | |  |
The allele frequencies in the different ethnic groups are summarized in [Table 1].
The commonest allele in the Sri Lankan population was the *3 null allele, which has no enzymatic activity (44.9%). This was most prevalent among the Sinhalese (60.3%). The second most common allele was the *10 allele, which confers reduced enzymatic activity (39.3%). This was most prevalent among the Moors (44%). The null allele *4 also occurred at a frequency of 12.2%. The difference in the frequency of the alleles between the three major ethnic groups in the Sri Lankan population was not significant. [Table 2] compares CYP2D6 allele frequencies among the Sri Lankans and some other populations.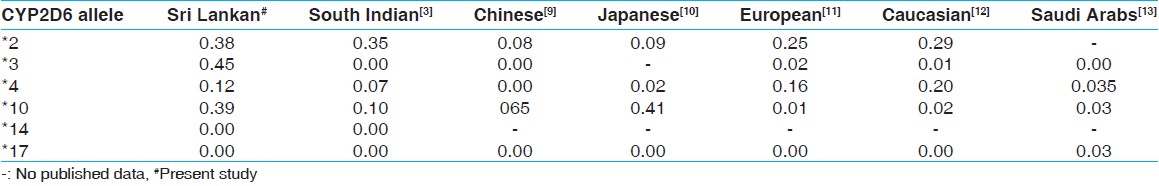 | Table 2: Comparison of CYP2D6 allelic frequencies in different population
Click here to view |
 Discussion Discussion | |  |
In this study, we set out to determine the prevalence of different alleles of the CYP2D6 gene in the Sri Lankan population. Since the number of CYP2D6 alleles is large we selected six key polymorphisms, which are most prominent in other major populations for analysis. One main observation made when comparing our results with those reported in a South Indian population was the high prevalence of *3 in all three ethnic groups in Sri Lanka, which is absent in the South Indian population. Even though there is a higher prevalence of enzyme inactivation alleles seen in the Sinhalese, phenotypically, the prevalence of PM phenotype is low (0-2%). [4] This is probably due to the heterogeneity of the CYP2D6 wild type. However, this warrants further studies with phenotypic-genotypic correlation in Sri Lankan subjects.
It is interesting to consider our results in the light of the different geographic origins of the major ethnic groups that form the Sri Lankan population. The Sinhalese are probably the descendants of the original inhabitants of the Island, but their origin is also traced to Northern India. However, there is disagreement as to whether they came originally from the North West or North East part of India. [14] A phenotypic study carried out in people living in Bombay, which is the North West part of India shows low prevalence (3%) of the PM phenotype. [15],[16] The Sri Lankan Tamils are the descendants of Tamils from South India who came to the Island and settled down at various times in the past when Sri Lanka was invaded by South Indian rulers. The Moors are the descendants of Arab traders. There is a higher prevalence of UM and EM alleles among the Arab population [17],[18] and the prevalence of null alleles is low. [13] The *3 allele is absent in South Indian Tamils, the presence of the *3 allele among a large proportion of Sri Lankan Tamils to the point where it is almost reaching the high frequency observed in Sinhalese probably suggests that there has been considerable admixture between these two populations following migration from South India. This is supported by the results of blood group genetic marker analysis, which also have not revealed any difference between the Sinhalese and Tamils. [12] This is further supported by the high prevalence of *2 and the absence of the *14 and *17 alleles in all the ethnic groups.
The *10 allele, considered an Asian specific allele, occurs in our population as frequently as in Japanese. [5],[10] However the presence of *3 allele, which is not reported in Asian populations and the presence of the Caucasian specific *4 allele in a higher frequency compared with other Asian populations [2],[6],[9],[13] are unique features of the frequency of these alleles in the Sri Lankan population found in this study. These findings are not surprising because Sri Lanka is an island nation, located at the center of the ancient naval route between the East and West, and thereby visited by many traveller, as well as invaded by Europeans from the 16 th century onwards resulting in gene admixture. Interestingly the absence of the African specific *17 allele in Sri Lankans suggest that there has been no gene admixture from populations of African descent. [8] This is again supported by the absence of significant African links with Sri Lankans in the past.
In summary, the most prevalent allele in the Sri Lankan population is the loss of function *3 allele, which is not present in the South Indian population. The *10 allele, which causes partial loss of enzyme activity was seen in 39% of Sri Lankans compared to only 10% in South Indians and the other loss of function allele *4, which is a Caucasian specific allele was found in 12% of our population. Therefore, a high percentage of Sri Lankans carry enzyme inactivation alleles of the CYP2D6 gene. This warrants prescription of smaller doses of drugs metabolized through CYP2D6 enzymes when such drugs are prescribed. This observation would be useful when formulating drug dosage guidelines for the Sri Lankan population. A phenotypic study of CYP2D6 enzyme activity in Sri Lankan population will further confirm these findings.
 Acknowledgment Acknowledgment | |  |
This study was funded by a grant from the Improving Relevance and Quality of Undergraduate Education Project, Faculty of Medicine, University of Colombo, Sri Lanka.
 References References | |  |
| 1. | Griese EU, Ilett KF, Kitteringham NR, Eichelbaum M, Powell H, Spargo RM, et al. Allele and genotype frequencies of polymorphic cytochromes P4502D6, 2C19 and 2E1 in aborigines from western Australia. Pharmacogenetics 2001;11:69-76. 
[PUBMED] |
| 2. | Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin Pharmacokinet 2009;48:689-723. 
[PUBMED] |
| 3. | Naveen AT, Adithan C, Soya SS, Gerard N, Krishnamoorthy R. CYP2D6 genetic polymorphism in South Indian populations. Biol Pharm Bull 2006;29:1655-8. 
[PUBMED] |
| 4. | Weerasuriya K, Jayakody RL, Smith CA, Wolf CR, Tucker GT, Lennard MS. Debrisoquine and mephenytoin oxidation in Sinhalese: A population study. Br J Clin Pharmacol 1994;38:466-70. 
[PUBMED] |
| 5. | Kirchheiner J, Brøsen K, Dahl ML, Gram LF, Kasper S, Roots I, et al. CYP2D6 and CYP2C19 genotype-based dose recommendations for antidepressants: A first step towards subpopulation-specific dosages. Acta Psychiatr Scand 2001;104:173-92. 
|
| 6. | Kubota T, Yamaura Y, Ohkawa N, Hara H, Chiba K. Frequencies of CYP2D6 mutant alleles in a normal Japanese population and metabolic activity of dextromethorphan O-demethylation in different CYP2D6 genotypes. Br J Clin Pharmacol 2000;50:31-4. 
[PUBMED] |
| 7. | Masimirembwa C, Persson I, Bertilsson L, Hasler J, Ingelman-Sundberg M. A novel mutant variant of the CYP2D6 gene (CYP2D6 *17) common in a black African population: Association with diminished debrisoquine hydroxylase activity. Br J Clin Pharmacol 1996;42:713-9. 
[PUBMED] |
| 8. | Gaedigk A, Bradford LD, Marcucci KA, Leeder JS. Unique CYP2D6 activity distribution and genotype-phenotype discordance in black Americans. Clin Pharmacol Ther 2002;72:76-89. 
[PUBMED] |
| 9. | Garcia-Barceló M, Chow LY, Chiu HF, Wing YK, Lee DT, Lam KL, et al. Genetic analysis of the CYP2D6 locus in a Hong Kong Chinese population. Clin Chem 2000;46:18-23. 
|
| 10. | Tateishi T, Chida M, Ariyoshi N, Mizorogi Y, Kamataki T, Kobayashi S. Analysis of the CYP2D6 gene in relation to dextromethorphan O-demethylation capacity in a Japanese population. Clin Pharmacol Ther 1999;65:570-5. 
[PUBMED] |
| 11. | Marez D, Legrand M, Sabbagh N, Lo Guidice JM, Spire C, Lafitte JJ, et al. Polymorphism of the cytochrome P450 CYP2D6 gene in a European population: Characterization of 48 mutations and 53 alleles, their frequencies and evolution. Pharmacogenetics 1997;7:193-202. 
[PUBMED] |
| 12. | Griese EU, Asante-Poku S, Ofori-Adjei D, Mikus G, Eichelbaum M. Analysis of the CYP2D6 gene mutations and their consequences for enzyme function in a West African population. Pharmacogenetics 1999;9:715-23. 
[PUBMED] |
| 13. | McLellan RA, Oscarson M, Seidegård J, Evans DA, Ingelman-Sundberg M. Frequent occurrence of CYP2D6 gene duplication in Saudi Arabians. Pharmacogenetics 1997;7:187-91. 
|
| 14. | Saha N. Blood genetic markers in Sri Lankan populations - Reappraisal of the legend of Prince Vijaya. Am J Phys Anthropol 1988;76:217-25. 
[PUBMED] |
| 15. | Lamba V, Lamba JK, Dilawari JB, Kohli KK. Genetic polymorphism of CYP2D6 in North Indian subjects. Eur J Clin Pharmacol 1998;54:787-91. 
[PUBMED] |
| 16. | Doshi BS, Kulkarni RD, Chauhan BL, Wilkinson GR. Frequency of impaired mephenytoin 4'- hydroxylation in an Indian population. Br J Clin Pharmacol 1990;30:779-80. 
[PUBMED] |
| 17. | Ingelman-Sundberg M. Duplication, multiduplication, and amplification of genes encoding drug-metabolizing enzymes: Evolutionary, toxicological, and clinical pharmacological aspects. Drug Metab Rev 1999;31:449-59. 
[PUBMED] |
| 18. | Islam SI, Idle JR, Smith RL. The polymorphic 4-hydroxylation of debrisoquine in a Saudi Arab population. Xenobiotica 1980;10:819-25. 
[PUBMED] |
[Table 1], [Table 2]
| This article has been cited by | | 1 |
Increased prevalence of functional minor allele variants of drug metabolizing CYP2B6 and CYP2D6 genes in Roma population samples |
|
| Agnes Weber,Renata Szalai,Csilla Sipeky,Lili Magyari,Marton Melegh,Luca Jaromi,Petra Matyas,Balazs Duga,Erzsebet Kovesdi,Kinga Hadzsiev,Bela Melegh | | Pharmacological Reports. 2014; | | [Pubmed] | [DOI] | |
|
 |
|