Sites of interest on the World Wide Web—edited by Rick Neubig and David Roman
Synaptic Plasticity and Glutamate Receptors
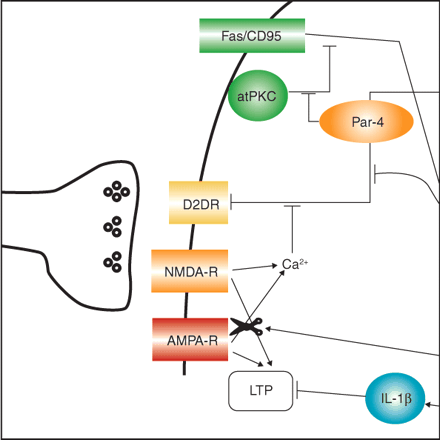
In this month’s MI, Mark Mattson and Marc Gleichmann highlight the unexpected role of Par-4 (prostate apoptosis response-4) in the regulation of neurotransmission at striatal dopaminergic synapses. The University of Bristol hosts the Medical Research Council Centre for Synaptic Plasticity (http://www.bris.ac.uk/Depts/Synaptic/), which is part of the MRC Cooperative on Neuronal Plasticity, Learning, and Memory. This Web site features many tools for researchers investigating aspects of synaptic plasticity—and for anyone interested in glutamate receptor function. The main portion of the site is divided into three major categories: 1) structure and function of glutamate receptors; 2) pharmacological tools; and 3) transgenic models. Selecting any of these areas brings focus to your search in a receptor-specific-manner, by AMPA, NMDA, kainate, and mGlu receptors. Each of the subtype-specific areas is filled with excellent graphical models and a wealth of information regarding modulation of these receptors and their roles in synaptic plasticity. The transgenic model page features referenced descriptions of phenotypes related to altered receptor expression, subdivided into sections describing global knockouts, tissue-specific knockouts, as well as expression of point mutants. The pharmacological tools section contains information about available agonists and antagonists and provides commentary about selectivity, Ki values, and tips for which ligands are best suited for discriminating among different receptor subtypes. This Web site provides a quick and usable guide for anyone looking more deeply into the complicated mechanisms of synaptic plasticity.
The WWW is Buzzing with Drosophila Resources
Also in this month’s MI, Alicia Celotto and Michael Palladino highlight the use of Drosophila as a model system for the study of neurodegeneration. The use of this tiny insect as a model system has advanced research in many fields for many decades. Now, there are a variety of resources on the Internet that catalog and organize the abundant amount of information known about these fascinating model organisms.
The Fly Brain Project (http://flybrain.neurobio.arizona.edu/Flybrain/html/index.html) provides a visible atlas to the Drosophila brain, relying on micrographs of brain slices, whole brain features, and 3D models of important neurological structures. The VRML (virtual reality modeling language) tours allow for the manipulation of brain models, providing flexibility for pinpointing specific brain areas. Furthermore, this Web site features a Genetic Dissection of the Brain as well as a Developmental Studies area with links to development models (as “movies”) and images of embryonic brain development. A pleasure to navigate, this site really allows the interested investigator to “get inside” the fly’s brain.
For practical needs, the Bloomington Drosophila Stock Center at Indiana University offers one-stop shopping for a vast collection of fly strains and genetic mutants—at a very informative Web site (http://fly.bio.indiana.edu/).
The WWW Virtual Fly Library (http://www.ceolas.org/fly/), hosted by Gerard Manning, serves as a great starting point for those searching for information about Drosophila. This site serves as a jumping-off point to many sites, both local and international, which provide a variety of information, pictures, models, and even fly husbandry tips.
- © American Society for Pharmacology and Experimental Theraputics 2005