Framework For Environmental Exposure Research: The Disease-First Approach
Major diseases confronting us today—so-called common diseases—are chronic and disabling conditions. These diseases are the largest cause of death worldwide, led by cardiovascular disease and followed by cancer, chronic lung diseases, and diabetes mellitus (1). The prevalence of common diseases is increasing. In developing countries, where common diseases have not displaced acute infectious disease, a protracted double burden of disease exists (2), and whereas public health practices have centered around infectious disease, the expanding public health burden from common diseases has been the subject of relatively modest attention.
It is clear that environmental factors, in combination with genetic factors, play a huge role in common disease susceptibility. Associations between environmental factors and health outcomes are, however, complex and poorly characterized (3). Levels of exposure, for example, are often difficult to ascertain, owing to a lack of detailed monitoring as well as to inevitable variations within any population. New technologies and genomic information, however, promise to meet this long-standing challenge. (See Figure 1⇓.) Common diseases require an understanding of gene–gene, gene–environment, and gene–vector–environment interactions. With the advent of the high-throughput techniques of genomics, proteomics, and metabolomics, biomedical science has been steadily shifting to a more comprehensive focus on understanding the diverse and complex responses underlying the development of disease. Coupled with increases in computational power and sophisticated informatics tools for data integration and modeling, researchers can develop quantitative models to support risk assessment.
Environmental (red) and genetic factors (blue) conspire to generate common disease (purple). Over time, environmental stressors (broadly defined, to include unhealthy behaviors as well as toxicants), along with genetic factors (such as metabolites and biomolecular activities), may act cumulatively to produce disease (left). But because disease course may be so variable (right), depending on a multiplicity of diverse factors, it has been impossible to assess the causative nature of many common chronic diseases. The availability of modern post-genomics methodologies now challenges environmental health science to detail disease causation in terms of all factors that fall under the rubrics of “environment” and “genetics.”
The current approaches to the exposure arm of exposure–disease relationships, which we term the “traditional approach” and the “hazard-assessment approach,” cannot generate the detailed information necessary for a comprehensive appreciation of common multifactorial diseases. These approaches, for example, have focused on chemical toxicants and other pollutants, usually in the general environment, such that they often cannot definitively link a specific substance to a common diseases. In partial response to this limitation, the working definition of “environment” for environmental health sciences has expanded to include a range of “real-world” factors that affect human health, including food, drink, and medicines, the behavioral choices we make, and the structures and infrastructure around us (the “built environment”) (4, 5). This broadened definition brings with it a new set of challenges, involving the difficulties of measuring the combined health effects of nutritional deficiencies, infections, pollutants, and behavioral stressors, each category of which represents a multiplicity of risks for disease. Clearly, assessments for these types of complex exposures will require new ways of thinking about environmental exposure analysis and assessment.
“Disease-First” Approach to Exposure Analysis
We propose here a systematic tracking of population health status that will allow us to obtain highly detailed information on combined exposures and provide measurable endpoints. The “disease-first” surveillance uses clinical analyses of population health status as the starting point for research inquiry. The assumption that health status reflects environmental exposures is not novel, of course, as public health practitioners and medical experts have made use of it for centuries [see (6)]. A truly comprehensive health status monitoring system for most common diseases in the US, however, is not yet in place, which is unfortunate because consistent and accurate health status data compiled over many decades could be used to reflect changes in environmental conditions. Infectious disease monitoring in the US and health status monitoring in Sweden are good examples of successful programs that illustrate how effective population monitoring can be.
Over the past five years, planning has been launched to extend existing efforts into a national capacity of health status monitoring in the US. An example is the announcement by the Department of Health and Human Services Secretary last year of a ten-year plan (“The Decade of Health Information Technology”) to transform the health care delivery system in the US by improving the health information infrastructure. This topic was also the focus of a recent symposium by the Institute of Medicine’s Roundtable on Environmental Health Sciences, Research, and Medicine (7), and is the focus of a new program of the Centers for Disease Control and Prevention (http://www.cdc.gov/nedss/index.htm), among other initiatives (8). Thus, the first component of the disease-first approach—the surveillance component—is gaining momentum.
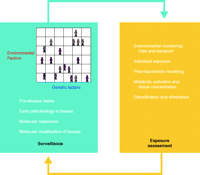
The second component of the disease-first approach aims to obtain a detailed molecular characterization of preclinical and clinical disease, and then to integrate this information with a vast body of information on environmental exposures, broadly defined. The approach starts with a specific common disease and detailed assessments of the molecular, cellular, and tissue changes that underlie pathogenesis in response to candidate environmental stressors. The disease-first approach will rely on the integration of information from multiple domains (Figure 2⇓), and especially on the blending of information from experimental models of disease and exposure analyses (9). This integration, which is the hallmark of the disease-first approach, is depicted in Figure 2⇓.
The disease-first approach to environmental health analysis of common disease. The two major components of the approach are highlighted. Integration of information (indicated by the arrows) from several specialties will depend on the development of informatics, experimental models of disease, and several other emerging technologies. See text for details.
In the disease-first approach, many different categories of information must be gathered and extensively integrated, including measurement of general atmospheric pollutants/toxicants, fate and transport of hazardous agents in the ecosystem and in the diet, human body burden measurements of agents, various endogeneous and xenobiotic metabolites, molecular markers of normal stress responses, and early pathophysiological changes that may precede disease onset (10–13). The disease-first hypothesis is that data amalgamated from these sectors will allow us to do a more sophisticated job of finding molecular signatures of exposures and linking them, through the power of informatics, with pathophysiological changes. In terms of pathophysiological processes, by the way, it is important to keep in mind that common diseases are manifestations of co-opted “robustness,” in which mechanisms that normally protect our bodies are effectively taken over to sustain and promote the disease states (14). Therefore, gaining a true understanding of common diseases will require an integrated perspective on the robustness of biological systems.
Research on dose–response relationships of candidate exposures using animal and cell models of common disease will be required to support a detailed disease-first approach. Studies with experimental models can assign molecular signatures to threshold exposure and delineate normal stress response from pathology. Knowledge of thresholds obtained in this way can be used to predict responses to new stressors and to understand how thresholds vary as a function of susceptibility factors.
Exposure Analyses
It is useful to review ongoing exposure analyses, because their maintenance and improvement are critically important. The “traditional approach” to the exposure arm of exposure–disease relationships, as reviewed elsewhere (15–17), can be conceptualized as a linear sequence. A typical sequence would begin with release of a chemical toxicant into the workplace and end with clinical evaluation of disease; the intermittent steps may include measurement of ambient exposures, personal exposures at the breathing zone, or biomonitoring of body fluids. Biotransformation, fate and transport, human uptake and metabolism, and target tissue dose are all part of this traditional and highly “researchable” framework (18–21). This framework is logical and has been used for years in the fields of exposure assessment and epidemiology. The experimental and computational methodologies used in the approach have become increasingly sophisticated and precise in recent years. Disadvantages of the approach, however, include difficulties in obtaining precise measurements of multifactorial or low-dose exposures and actual internal human doses.
The “hazard-assessment” approach, like the traditional approach, starts with a suspected hazardous agent. Through use of a range of laboratory methodologies, this approach characterizes relationships between dose of an agent and response in a model organism. The work of the US National Toxicology Program (http://ntp.niehs.nih.gov/) is one of many examples of large-scale research in this area. This approach, however, is capable of providing only a piece (hazard identification) of the larger puzzle of understanding the health effects of exposure in the general population.
Finally, in academic research, it has become obvious that neither of these approaches (traditional and hazard-assessment) is sufficiently “hypothesis-driven” and robust to attract the large funding commitments necessary to investigate the exposure arm of gene–environment interactions. Thus, finding additional avenues to fill the information gaps on exposure has been a practical challenge for the field of environmental health research.
New Tools for a New Approach
A proposal to expand exposure research to include tools for correlating exposure, tissue response, and molecular characterization of common disease would have been premature five years ago. However, the measurement and computational tools necessary to obtain data corresponding to the surveillance arm of the “disease-first” approach will soon be available. New high-throughput analytical methodologies and the various “genomics” disciplines are maturing rapidly, their application is becoming more cost-effective, and they will be used widely in medical therapeutics within the next five years (22, 23). Bioinformatics is also progressing toward meeting the challenge of extracting information from the mountains of data generated by these new technologies. The use of improved animal models of disease will facilitate more precise experimentation and reveal more comprehensive insight into mammalian physiology and pathophysiology, as will recent research innovations such as RNA interference and subcellular imaging. In addition, national and regional mapping efforts by geospatial information systems are expanding and beginning to revolutionize the area of exposure assessment (24). Sensor-based technologies (e.g., nano-technology) will soon contribute new capabilities in general environmental monitoring of toxicants and pollutants as well as in individual monitoring. Such indicators as dietary patterns, physical activity patterns, and behavioral and lifestyle patterns will be accessible on a cost-effective basis.
The Disease-First Approach in Action: Liver Cancer and Parkinson’s Disease
One of the best examples of success with a disease-first approach is the case of research on liver cancer (hepato-cellular carcinoma) in China and Taiwan, beginning in the 1960s, that established that risk for disease varied as a function of dietary ingestion of aflatoxins in combination with hepatis B virus infection. This success story, involving several decades of research, started with disease surveillance and early epidemiology studies in the 1960s and 1970s. Careful detective work by researchers identified candidate agents in the environment, and eventually more extensive population-based studies were conducted to assess the candidate agents. The scientific inquiry eventually involved a range of laboratory-based studies on metabolism of the primary harmful agent, aflatoxin B1, and on aflatoxin-induced molecular changes in DNA and in liver oncoproteins such as p53. Finally, molecular biomarkers developed in these studies were used in large population-based studies. The information produced by this research has had a large impact in prevention of liver cancer, as reviewed by Groopman and colleagues (16).
Another example is emerging from “environmental” approaches to Parkinson’s disease (PD). This story exemplifies the integrative work of laboratory researchers, population geneticists, clinicians, and epidemiologists. Clinical observations in the 1980s led to the recognition that 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) was a causative factor in a PD-like syndrome. MPTP, in turn, led to animal models of PD, opened lines of research on other candidate environmental exposures, and provided a framework for studying nigral cell death mechanisms (25–31). The identification of genetic susceptibilities to PD has enhanced understanding of the molecular etiology of the disease and indicated how environmental exposures might trigger disease. Studies of familial PD have also revealed several disease susceptibility genes, including alpha-synuclein and parkin, as have gene and allelic association studies (32–34). The candidate genes have been those with suspected mechanisms of PD pathogenesis; a number of genetic associations with PD have been reported for genes involved in detoxification of toxicants (e.g., CYP2D6 and MAOB). The recognition that many toxicants are oxidative stressors (35–37) also suggests candidate genes to be investigated with regard to PD.
Finally, many collaborative, multidisciplinary research efforts organized as disease-first initiatives in exposure–disease relationships are already underway. Recent examples supported by the National Institutes of Health include a PD research consortium (Collaborative Centers for Parkinson’s Disease Environmental Research), the Breast Cancer and the Environment Research Centers, and others.
The Challenge
The establishment of a national health status monitoring capacity will be a major long-term effort (see Figure 2⇑). But such a “big science” effort is clearly worth pursuing. The public has been willing to support such efforts in environmental health in the past when the objectives justified cost. As Francis Collins noted when the Human Genome Project was nearing completion, “[This is] the end of the beginning…the true payoff from the Human Genome Project will be the ability to diagnose, treat, and prevent disease, and most of those benefits to humanity still lie ahead.” Indeed, Collins (38), John Potter at the University of Washington (39), and others have suggested that the next steps should include large-scale, long-term, population-based research to investigate gene–environment interactions. Several efforts comprising large population-based studies have been proposed worldwide, such as the American Genes and Environment Study and the National Children’s Study in the US. Clearly, the time is right to accept the challenge of implementing the big science represented by the disease-first approach; it promises benefits that will justify the cost.
Acknowledgments
We thank Ernest Hood for editorial assistance and Shirley Isenhour for assistance with preparing the manuscript. We thank Tina Bahadori, David Brown, Allen Dearry, Judy Graham, John Groopman, Carol Henry, and Paul Lioy for comments on the manuscript. Submitted March 2, 2005.
- © American Society for Pharmacology and Experimental Theraputics 2005
References

Samuel H. Wilson, MD, is Deputy Director of the National Institute of Environmental Health Sciences, National Institutes of Health. Address correspondence to SHW. E-mail wilson5{at}niehs.nih.gov.

William A. Suk, PhD, is Director of the NIEHS Superfund Basic Research Program.